ChroMap - Data files / formats
ChroMap can display a variety of different data types.
Different views may be used to visualise these data types:
- Ideogram
- Gene info
- CGH-data
- Gene expression profiling data
You may use any kind of data to be displayed in
ChroMap, provided you bring your data into the form and structure of one of the data
ChroMap expects.
Ideogram
Gives a sketch of the chromosome banding pattern after Giemsa staining of metaphase chromsomes (g-banding).
A Recent (but not regularly updated) version of the ideogram file may be downloaded from ChroMap Site:
- homo sapiens
The ideogram data file shold look like:
| #chromosome | arm | band | iscn_start | iscn_stop | bp_start | bp_stop | stain | density | bases |
| 1 | p | ter | 0 | 1 | 0 | 0 | | | 1 |
| 1 | p | | 0 | 7335 | 1 | 124300000 | | | 124300000 |
| 1 | p | 3 | 0 | 4852 | 1 | 84700000 | | | 84700000 |
| 1 | p | 36 | 0 | 1521 | 1 | 27800000 | | | 27800000 |
| 1 | p | 36.3 | 0 | 344 | 1 | 7100000 | | | 7100000 |
| 1 | p | 36.33 | 0 | 100 | 1 | 2300000 | gneg | | 2300000 |
| 1 | p | 11.1a | 7282 | 7335 | 123500001 | 124300000 | acen | | 800000 |
| 1 | cen | | 7335 | 7335 | 124300000 | 124300000 | | | 1 |
| 1 | p | 10 | 7335 | 7335 | 124300000 | 124300000 | | | 1 |
| 1 | q | | 7335 | 15100 | 124300000 | 247249719 | | | 122949720 |
Gene info
With this data type you may show gene information next to the chromosomes.
ChroMap expects as original source for the gene information NCBI's seq_gene.md file.
To get a most recent version of these data, visit NCBI's ftp site:
Change to folder e.g. "genomes/MapView/Homo_sapiens/sequence/BUILD.37.2/initial_release/"
(use always highest BUILD.xxxx to get most recent version).
Or try folder "ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/mapview/"
Download the "seq_gene.md.gz" file, unpack the GZipped file (e.g. with free 7Zip tool).
The data file looks like:
| #tax_id | chromosome | chr_start | chr_stop | chr_orient | contig | ctg_start | ctg_stop | ctg_orient | feature_name | feature_id | feature_type | group_label | transcript | evidence_code |
| 9606 | 1 | 815 | 19836 | - | NT_077402.1 | 815 | 19836 | - | LOC653635 | GeneID:653635 | GENE | reference | - | mRNA;; |
| 9606 | 1 | 910 | 13214 | + | NW_921350.1 | 910 | 13214 | + | LOC642097 | GeneID:642097 | GENE | Celera | - | protein;identical;N |
ChroMap uses the columns:
- Chromosome
- chr_start
- chr_stop
- chr_orient
- feature_name
- feature_type
Now you can import the Gene-Info file into ChroMap.
Select from ChroMap Main menu | Utilities | Data files | Import | Gene list:
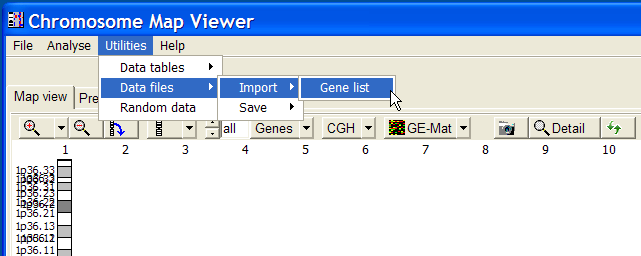
In the file selection dialog, select the just downloaded and un-zipped seq-gene.md file.
Importing of the file may take a while, as it may contain hundred thousands of gene features
To speed up loading of the gene info next time, save the file in a binary format.
Select Main menu | Utilities | Data files | Save | Gene list:
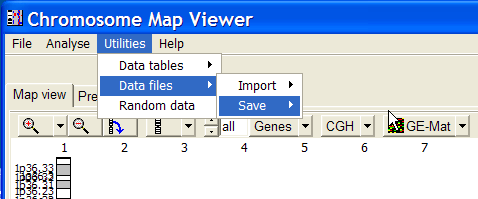
Save the filder under the name "GeneList.cfl" in a "data" folder under ChroMap's program frolder (e.g. " c:\programs\chromap\data").
Next time you start ChroMap, it will automatically load the new generated gene-info file.
The original "seq_gene.md" file from NCBI may contains somewhat redundant information.
The file contains informations from:
- different genome builds
- gene
- mRNA
- pseudo genes
- ...
To speed up data loading, it may be helpful to filter the original file.
Keep only those infos you would like to see in you analyses, and remove all other.
We use TableButler's Row-Filter to perform such tasks.
A Recent (but not regularly updated) binary gene infor file may be download from ChroMap Site:
- Complete list. This file contains all informations provided by NCBI (~1.5 million features, ~130 MByte).
- "Gene" list. This file contains only "Gene" informations provided by NCBI (~120.000 features ~10 MByte).
- "Gene - GRC" list. This file contains only "Genes" from the Genome Reference Consortium's Assembly Build 37 (~40.000 features, ~4 MByte). Celera Build 2001 as well as CAG-Toronto Build 2004 are removed.
Download either of the two files. Rename it to "GeneList.cfl" and place it in a "data" folder below ChroMap's program file location.
If you want to display other information then localisation of genes within ChroMap, bring your data into NCBI's "seq_gene.md" format and import it.
CGH-data
Gene expression profiling data

