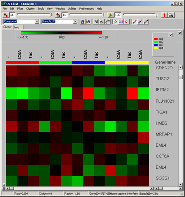
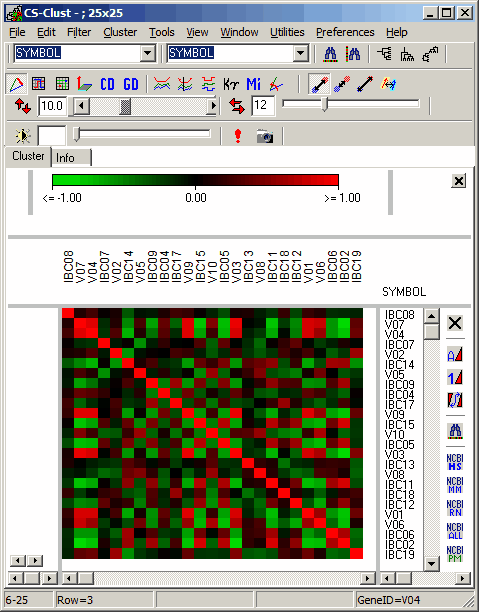
visualising intensities / ratios
of the expression matrix.
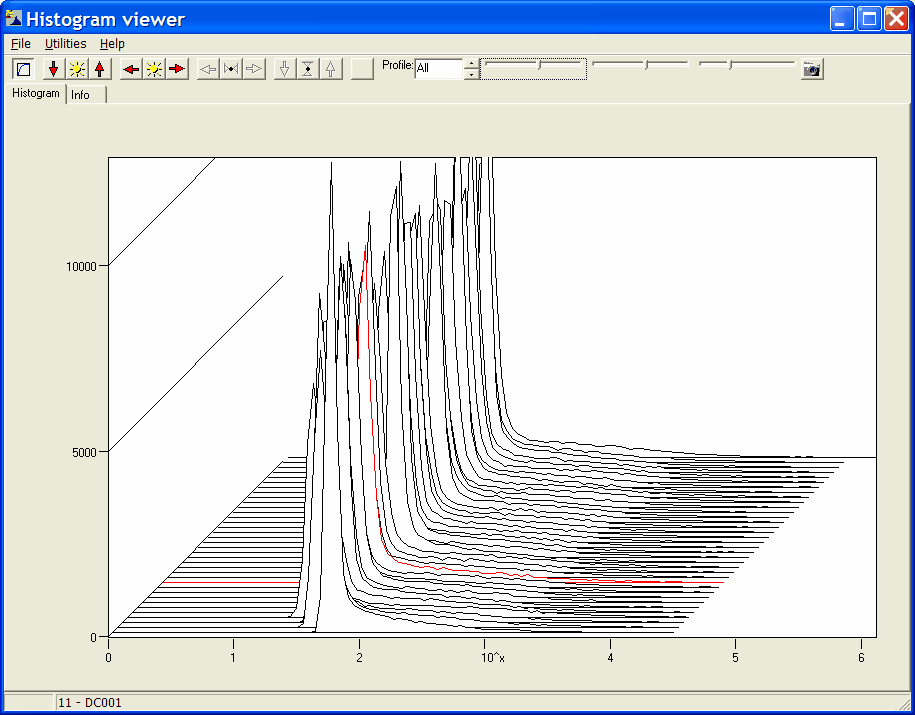
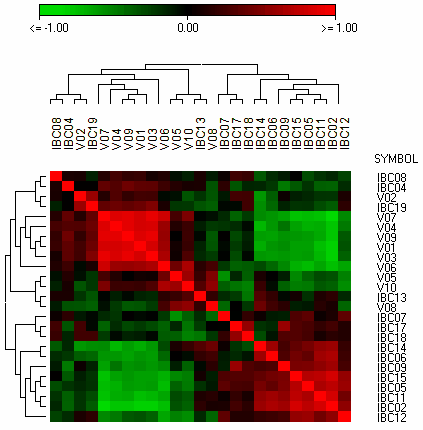
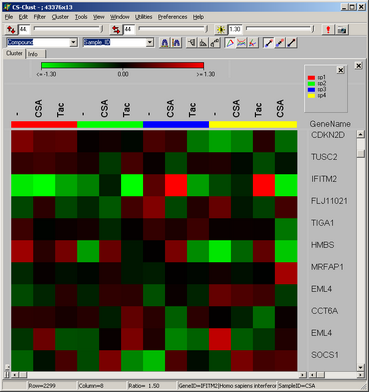
The famous red-green heat maps
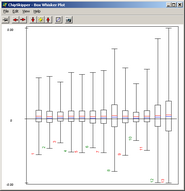
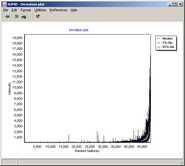
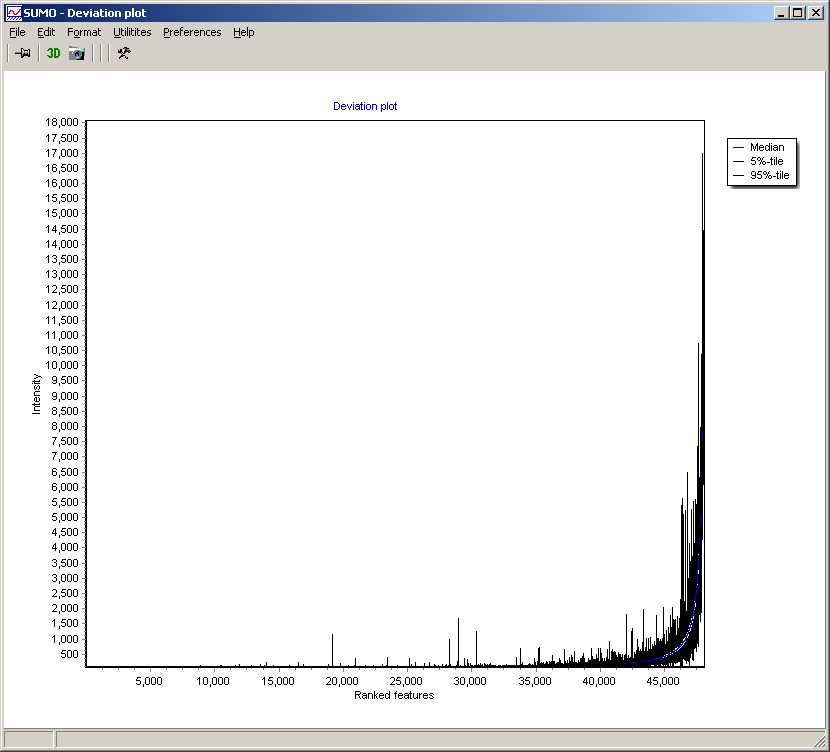
summarizing global intensity/ratio distribution
in all hybridisations
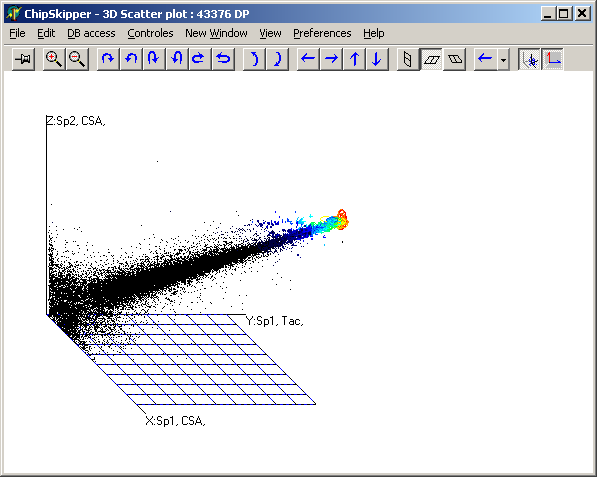
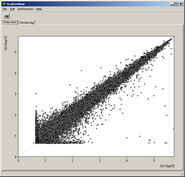
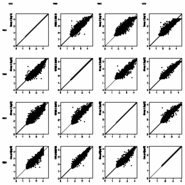
Analyze pair wise signal distributions
See more details about the scatterplot viewer.
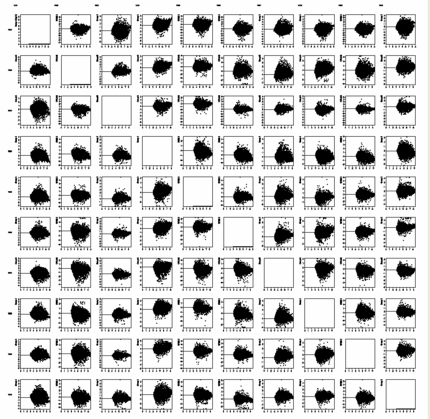
of hybridisations
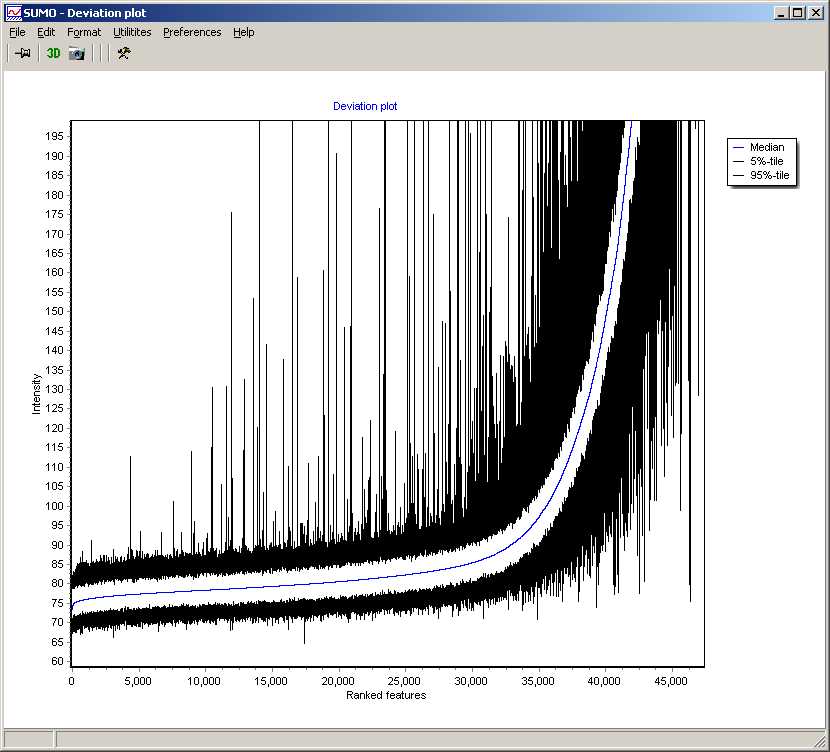
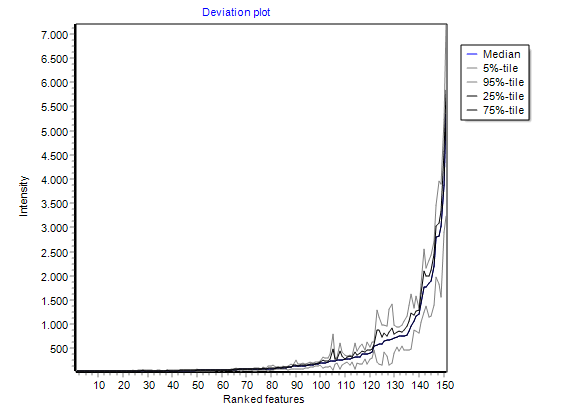
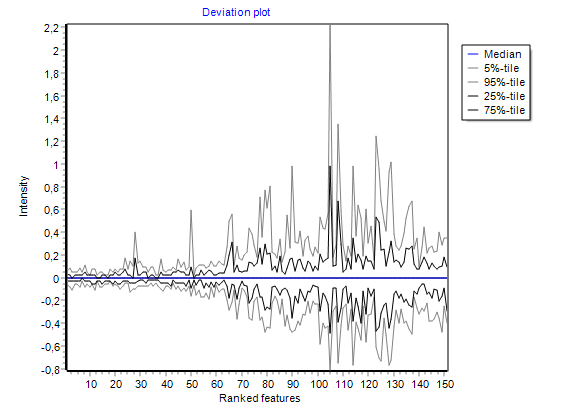
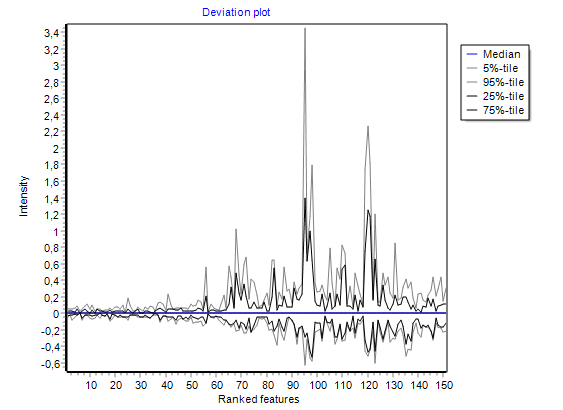
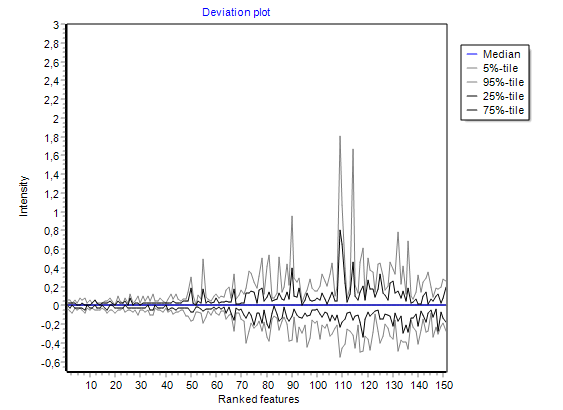
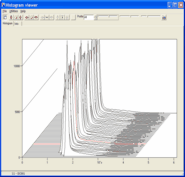
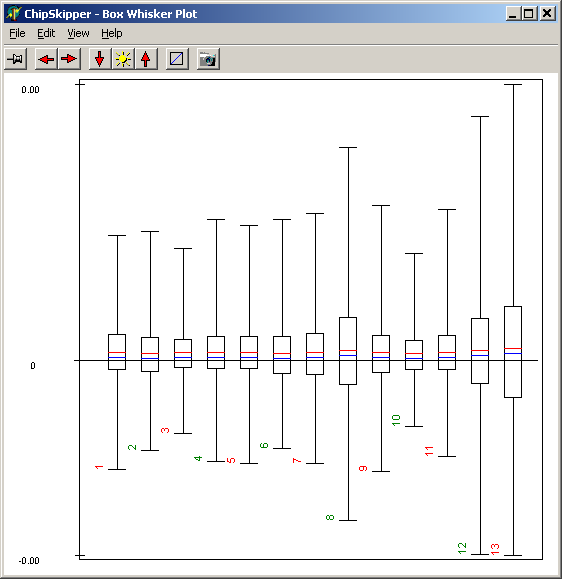
Get an overview of signal distributions
in all your sample data
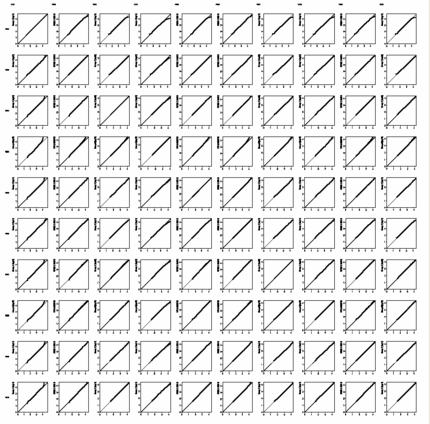
Show signal distribution in any
or all loaded samples
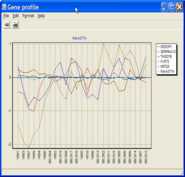
Illustrating e.g. gene/sample
data profiles
Illustrating inter-sample
similarity
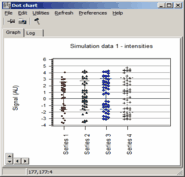
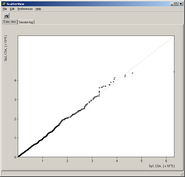
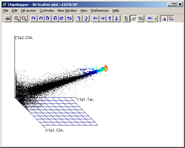
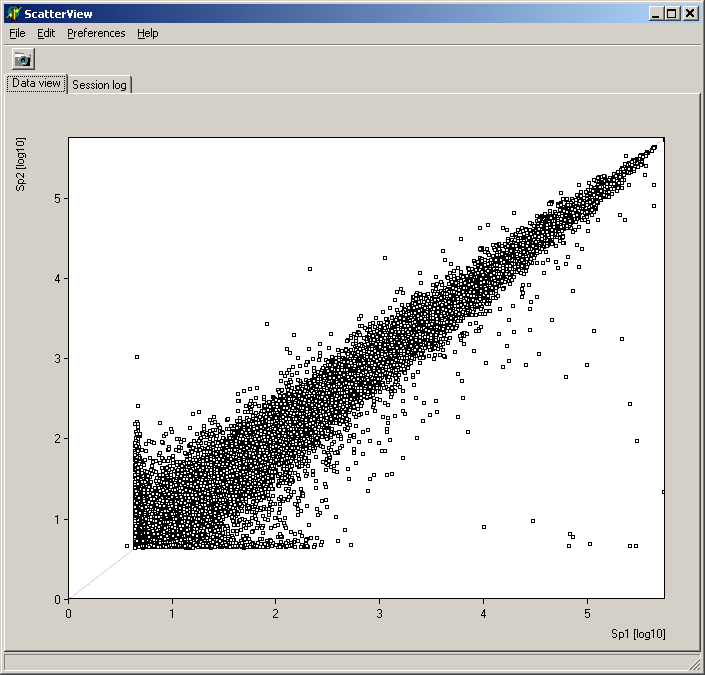
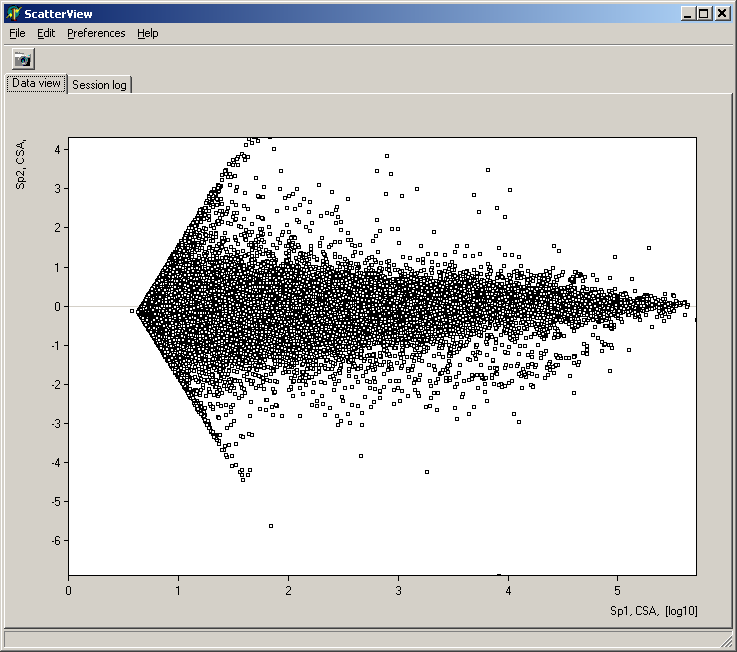
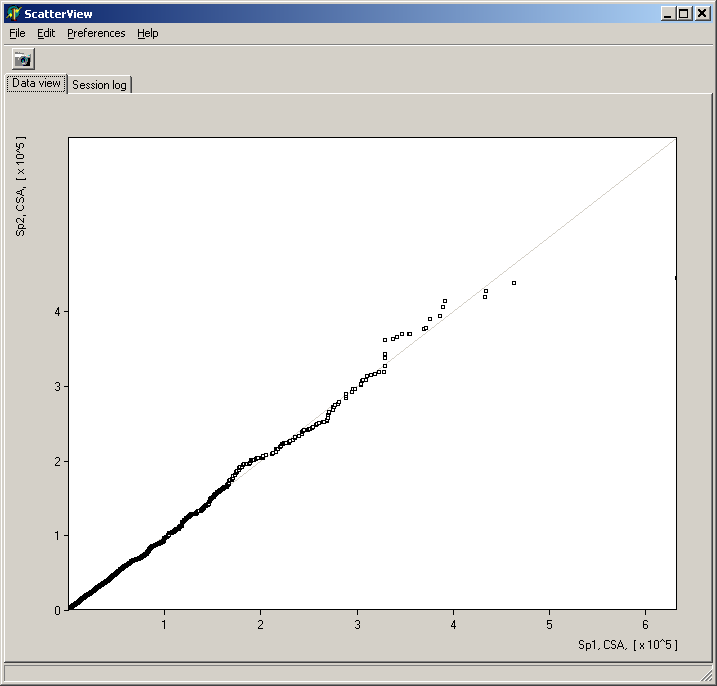 See more details about the Scatterplot viewer
See more details about the Scatterplot viewer