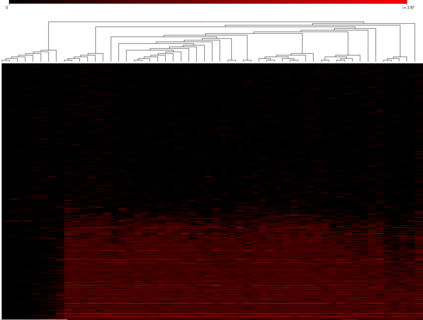
...
chr8 66429274 66429274 C T exonic RRS1 . nonsynonymous SNV RRS1:NM_015169:exon1:c.C143T:p.T48I 8q13.1 ...
chr8 66429276 66429276 G A exonic RRS1 . nonsynonymous SNV RRS1:NM_015169:exon1:c.G145A:p.G49R 8q13.1 ...
chr8 66429282 66429282 C T exonic RRS1 . nonsynonymous SNV RRS1:NM_015169:exon1:c.C151T:p.R51W 8q13.1 ...
...
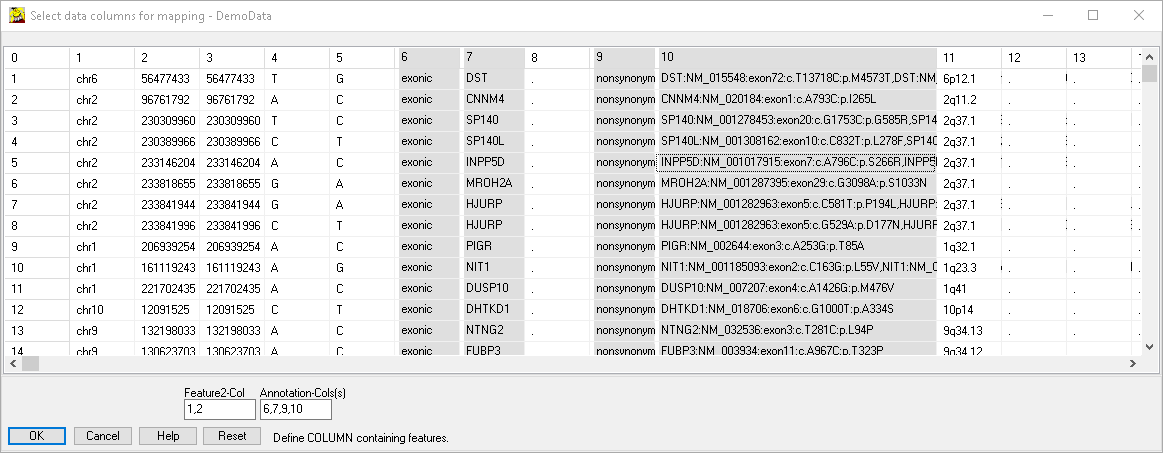
Keys Anno1 Anno2 Anno3 Total-Count AS-412869.vcf AS-412870.vcf _AS-412871.vcf ...
Total-counts -- -- -- -- 108 817 1504 3070 ...
...
chr1_100007135 SLC35A3 SLC35A3:NM_001271684:exon4:c.G444A:p.M148I 1p21.2 2 0 0 0 0 ...
chr1_102965516 COL11A1 COL11A1:NM_080630:exon36:c.C2539T:p.P847S 1p21.1 1 0 0 0 0
chr1_103754590 AMY1B AMY1B:NM_001008218:exon7:c.G970A:p.G324R 1p21.1 1 0 0 0 1 ...
chr1_1043688 AGRN AGRN:NM_198576:exon9:c.G1754T:p.C585F 1p36.33 3 0 0 0 1 ...
chr1_1044023 AGRN AGRN:NM_198576:exon10:c.G1999A:p.A667T 1p36.33 1 0 0 1 1 ...
chr1_10653830 CASZ1 CASZ1:NM_001079843:exon11:c.A2227G:p.T743A 1p36.22 1 0 0 0 1 ...
...