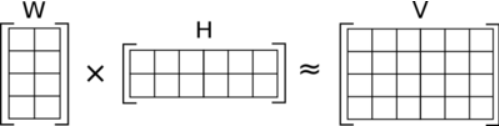
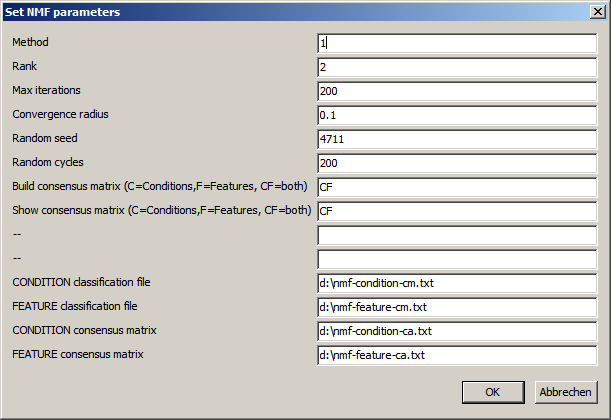
| Method | Define how the iterative is performed. 1=Lee, Seung: Multiplicative update with Educledean distance as cost function 2=Lee, Seung: Multiplicative update with Kullback-Leibler divergence as cost function ... |
| Rank | Number of factors for Conditions/features Should be a small number (2..10..) Give a range (e.g. "2,5") to run 4 complete rounds of NMF. Starting with NMF rank=2, followed by NMF rank=3, .... Result files are generated with a tag defining the rank (e.g. *_r2.txt for all results from NMF with rank=2) |
| Max. iterations | Maximumn number of iterations to perform in a single NMF run In case the NMF problem can not be solved the process might run infinitely. After user defned maximumn number of iteratons the update process is interrupted independent of convergence. |
| Convergence radius | After each update cycle, the relative change of the cost function is computed. The cost function should assymptotically approach Zero (but probably never reach it). Define the maximal accepted change (in %) of the cost funcion to stop the updating, harvesting a close approximation to a solution of NMF. As smaller the convergence radus as better the approximated solution, and as more update loops are required |
| Random seed | W and H are initialized with random numbers. Define a fixed start value to ensure the same sequence of random numbers. This may be hepful, to compare diffrent NMF runs with varying parameters. Leave the field empty for completely random sequences. |
| Random cycles | Number of NMF cycles with different randomly initialized W and H matrices. Condition/Feature associations to factors are collected and will be used to build a consensus similarity matrix. |
| Build consensus matrix C: only for the conditons F: only for features CF: both empty: none |
Factor asignments for condtitons/features are collected after each random cylce and
saved in the condition-/feature factor file. Additionally you may build the consensus similartity matrix and save it (condition & feature consensus matrix). Building of consensus matrices becomes time consuming (dim > 10000) and requires a lot of memory which may crash the program (dim > 35000). Thus, you may only build one (the smaller) matrix, or build the matrix later, independently reusing the condition-/feature factor files. |
| Show consensus matrix C: only for the conditons F: only for features CF: both empty: none |
Having build the consensus similarity matrix, you may immediately show data in a heatmap viewer. With matrices dim > 20000 this may crash SUMO. |
| Files | Base names for result files. SUMO will use the base names to store results from the different randomization rounds at the specified location. (e.g. "d:\nmf_Condition_cm_r1.txt" as consensus similarity matrix for ramdom round 1. |
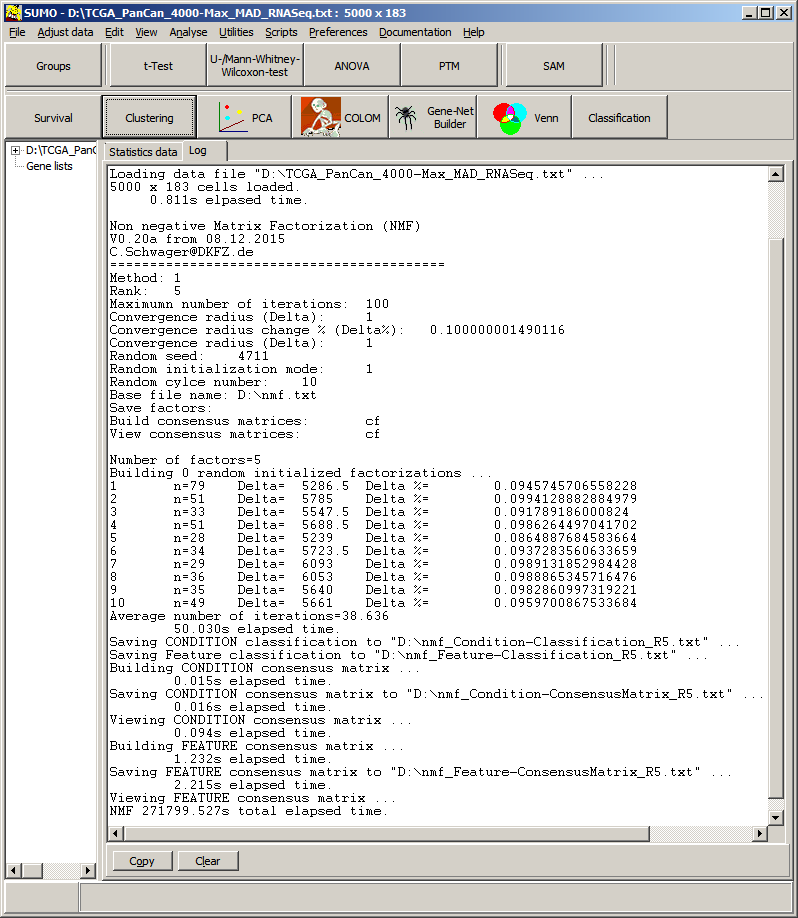
| # | Number of random round (e.g. 1=first round) |
| n | Number of iterations in this round. (e.g. 94 iterations for round 1, if n equals user defined number of max iterations, NMF didn't reach user defined accuracy. May be adjust parameters and run again. |
| Delta | Value of cost functions at last performed round. Delta=abs(Costfunction[i-1]-Costfunction[i]; |
| Delta % | Relative change of cost function between secod last and last random round. Delta%=(Delta / CostFunction[i-1])*100; Here, cost function changed between last two random rounds by ~0.097%. This is smaller user defined convergence radius => stop iterative update scheme and finalize randoum round 1. |
| Features (Rows) |
Conditions (Columns) |
t NMF (s) | Condition matrix t (s) |
Save C-CM t (s) |
Feature matrix t (s) |
Save F-CM t (s) |
Total t (s) | |
|---|---|---|---|---|---|---|---|---|
| 100 | 100 | 120 | 0.047 | 0.015 | 0.187 | 0.016 | 0.016 | 14.820 |
| 200 | 100 | 105 | 24.134 | 0.047 | 0.015 | 0.016 | 0.016 | 24.695 |
| 500 | 100 | 101 | 55.458 | 0.047 | 0.016 | 0.031 | 0.047 | 56.004 |
| 1000 | 100 | 106 | 115.113 | 0.063 | 0.015 | 0.109 | 0.156 | 115.878 |
| 2000 | 100 | 107 | 231.568 | 0.063 | 0.016 | 0.873 | 0.578 | 233.643 |
| 5000 | 100 | 104 | 565.020 | 0.109 | 0.015 | 6.489 | 3.526 | 576.080 |
| 10000 | 100 | 103 | 1432.183 | 0.172 | 0.015 | 30.763 | 14.337 | 1480.060 |
| 15000 | 100 | 104 | 2094.563 | 0.219 | 0.015 | 75.255 | 31.887 | 2207.351 |