For many tests, groups with different phenotypes have to
be defined.
E.g. distinguish between different cancer types. Therefore, those hybridisations
which are generated from the different cancer types (e.g. liver, kidney, lung,
brain, ..) have to be defined.
Click the Groups button:

The group selection dialog pops-up:
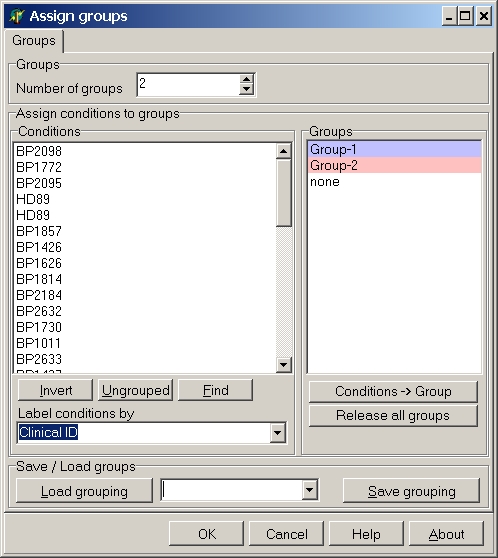
Depending on the analysis 1 or more groups have to be defined.
Set the number of groups by clicking the up/down buttons
or the Number of groups edit field or type
in the required number (in the example 2).
In the Groups list the corresponding groups are shown:
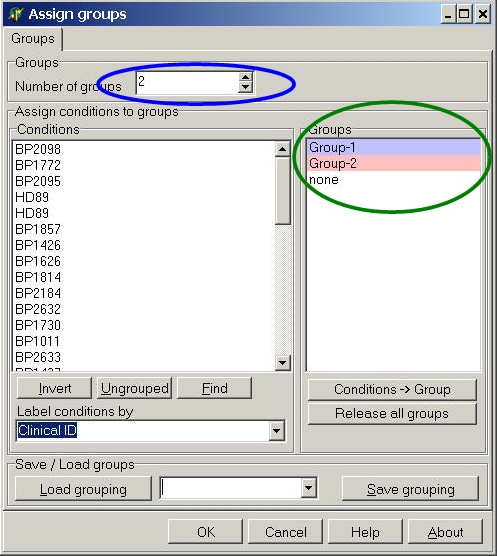
CS-SPAM sets the default group numbers according to the selected analysis:
In the Conditions list select the desired hybridisations.
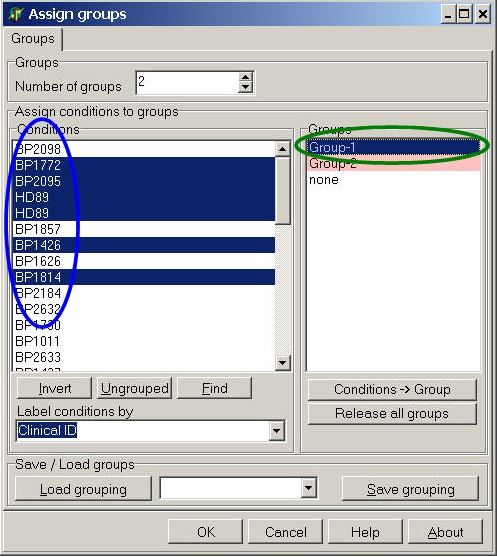
To select multiple conditions use Shift+Left mouse or
Ctrl+Left mouse.
Now select the corresponding group in Groups
list.
Double-click this group or click Conditions->Group button.
All selected conditions are now member of this group and get the corresponding colour:
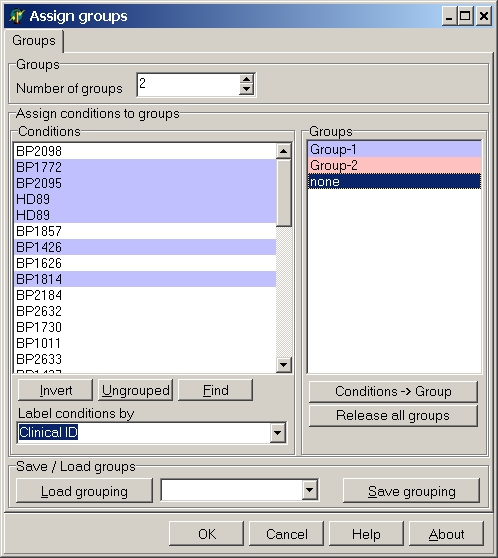
Do the same for the other group(s).
Click right mouse button in the group list:
From the context menu select:
If there are multiple header rows in a expression data matrix, the different rows may be used for selection of groups.
Open the Labels conditions by drop down list:
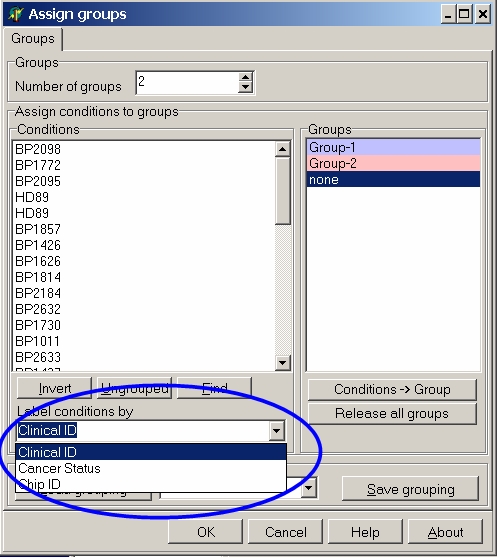
In the example, there were three header rows in the expression matrix file:
If you want to group the hybridisations by Cancer status select Cancer status from the Label conditions by list:
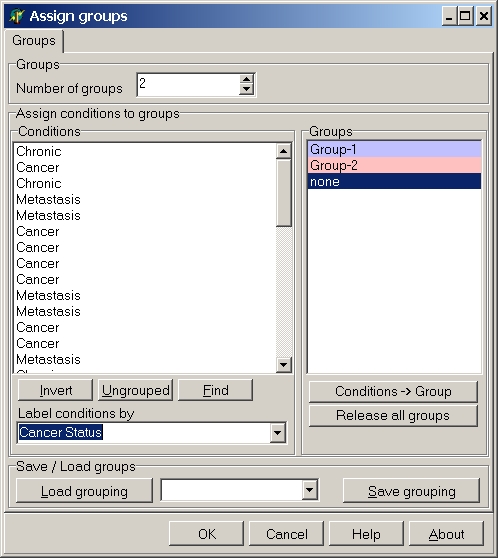
Now you can easily find those hybs with the same cancer status.
More easily, CS-SPAM can search and find conditions with similar naming.
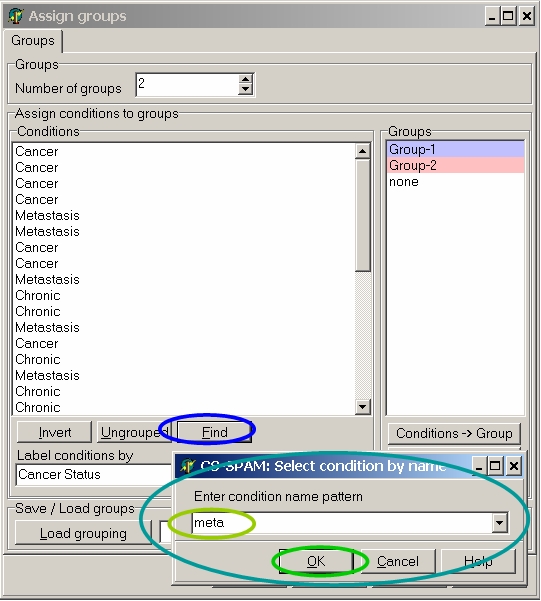
Click the Find button:
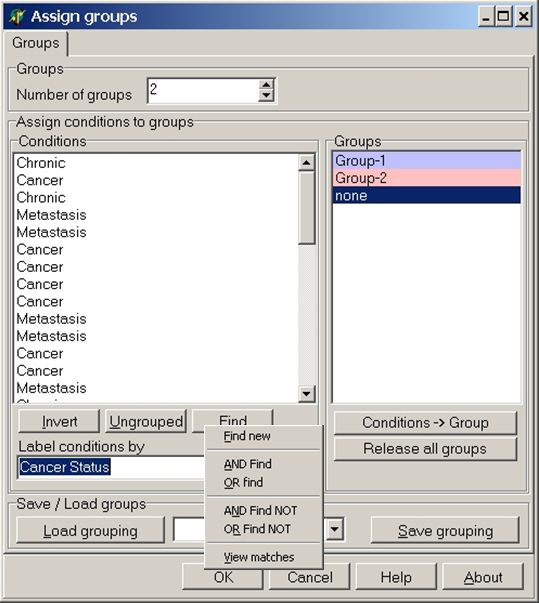
From the context menu pops-up select
In the input dialog type "meta" and click OK button to select all hybs which contain "meta" in their name:
All hybs which contain "meta" in their name are selected and can be assigned to a group:
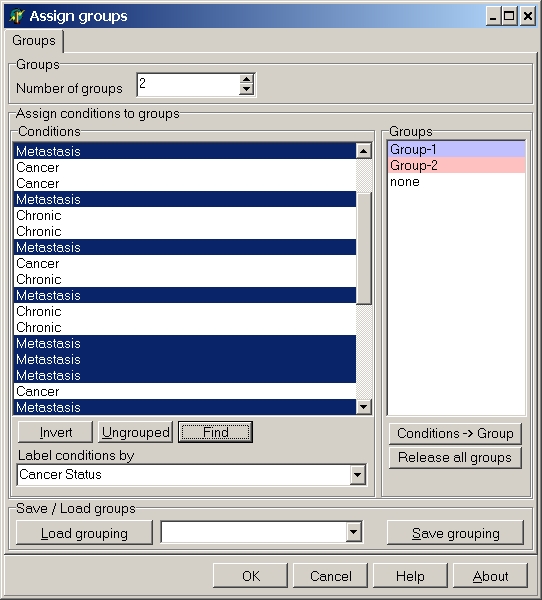
Similarly you can select the "Cancer" and "Chronic" hybridisations.
If you want to select both Cancer and Metastasis (logically the key should contain either Cancer or Metastasis) type
"can meta"
separated by SPACE.
Sometimes it may be useful to reuse a partial selection.
Assume you have hybs from different normal, cancer, metastasis, ... tissues from
different organs (lung, liver, kidney, brain, ...) which have been treated with
different drugs, and radiation therapies.
Thus is may be useful to build a selection for all cancer hybs which have not
been chemically treated. Now build subgroups from these for the different
radiation treatments.
To do this, build a selection.
Click right mouse button on the conditions list:
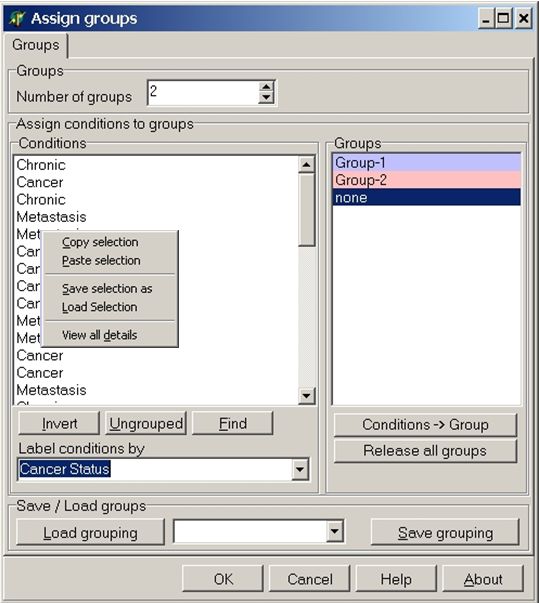
From the context menu select:
If two groups are required and all hybridisations shall be used it is enough to select and assign the hybs for group one.
Then click the Invert button to unmark the selected hybs and mark the up-to-now unselected hybs and assign those to group 2.
If multiple groups are required an all hybs shall be used, you can define the last group (all hybs which have not bee assigned to groups yet) by clicking the Ungrouped button.
If you want to reuse the grouping in another CS-Spam session, click the Save grouping button and save the selection list as simple text file.
To reuse a previously saved grouping, click the Load grouping button. and select a previously saved groupings text file.
The drop-down list contains the names of recent used group files
last edited 29.08.2006,