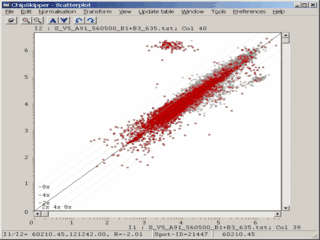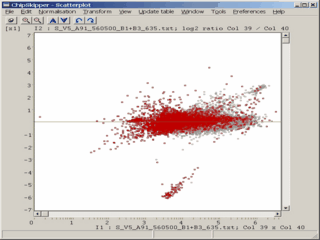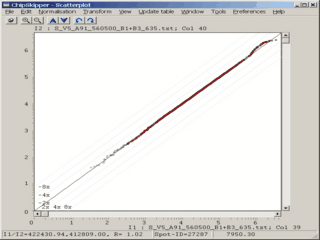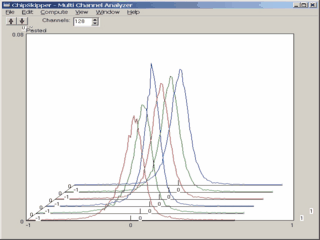
When analyzing results from microarray experiments we encounter always similar problems :
All this should happen with result files from our genome-wide yeast,
mouse
and human
cDNA chips ranging from 14000 to 55000 features. Sometimes Filter-arrays with up to 200,000 features
are used.
Some published data sets contain several hundred of data columns
Obvious solution might be to try MS Excel for these tasks. But this showed to become very time consuming, boring and error prone. Especially use of differing national settings (currency, date, decimal divider) may result in really nasty things happening automatically to your data. On the other hand Excel tends to crash when handling huge tables. Large data files (>65530 rows or >255 columns) can not be loaded completely anyhow.
Therefore, we implemented a tool which can do all the above described tasks (and more) : It serves us the nicely rearranged data tables like a butler : TableButler.
The program works on tab delimited files. In "batch mode" dozens (tested) or hundreds of files - even all hybridisation files on your hard disk - might be selected and processed in one go.
All tasks are configured interactively. The parameters for each task may be saved and recalled later.
A simple scripting
language allows to set up and store complete processing pipelines. Scripts can
be loaded and executed manually. The name of a script can be passed to TableButler
via a command parameter, thus a Desktop Icon can be used to start a TableButler
processing.
Furthermore, TableButler can be run as a server:
Several directories may be watched. Any TableButler script dropped
into the watched directories is automatically loaded and processed.
You can modify TableButler's appearance according to your preferences using command line parameters or ini-files
All functions may be addressed and configured from a graphical user interface. Previews of source and result files facilitate set up of filters and parameters with a view mouse clicks. Result files may replace the originals or saved separately.
For more details go here.