| "\x≥nnnn.nn" | all objects where X-valiue is equal or larger specified threshold |
| "\x≤nnnn.nn" | all objects where X-valiue is equal or smaller specified threshold |
| "\y≥nnnn.nn" | all objects where Y-valiue is equal or larger specified threshold |
| "\y≤nnnn.nn" | all objects where Y-valiue is equal or smaller specified threshold |
| "\s≥nnnn.nn" | all objects where size-value is equal or larger specified threshold |
| "\s≤nnnn.nn" | all objects where size-value is equal or smaller specified threshold |
| "\c≥nnnn.nn" | all objects where color-value is equal or larger specified threshold |
| "\c≤nnnn.nn" | all objects where color-value is equal or smaller specified threshold |
| "\r≥nnnn.nn" | all objects where RATIO x/y≥specified threshold |
| "\r≤nnnn.nn" | all objects where RATIO x/y≤specified threshold |
| "\lr≥nnnn.nn" | all objects where RATIO log2(x/y)≥specified threshold |
| "\lr≤nnnn.nn" | all objects where RATIO log2(x/y)≤specified threshold |
X Y Error-X Error-Y Size Color Group Name 128546.48438 115674.15625 1820.84790 11942.00098 34.01107 39.77178 0 1 30870.58594 133569.12500 2894.34741 13599.37695 3.44957 9.55785 0 2 -41971.60938 -54666.37500 3154.14136 5431.87061 6.53209 3.38831 0 3 -34792.36719 -68183.10938 3170.89355 2756.95752 58.64328 63.78699 0 4 -18017.51563 -98872.61719 1449.07239 8313.46094 25.48250 19.61693 0 5 96628.62500 42455.34375 576.08539 3467.49487 2.25695 75.79591 0 6 ...
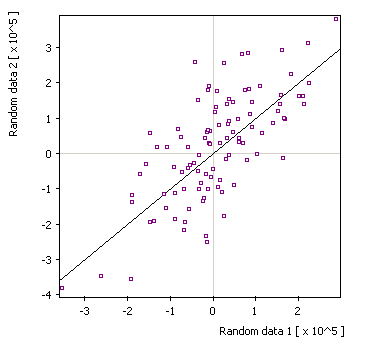
Default display: |
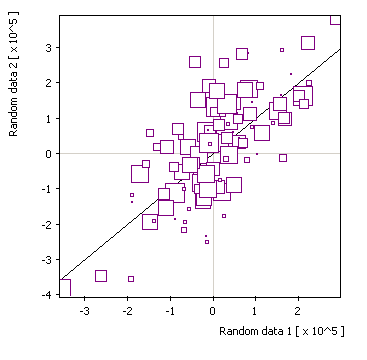
x/y + size: |
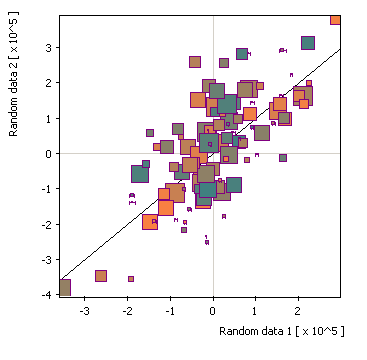
x/y + size/color: |
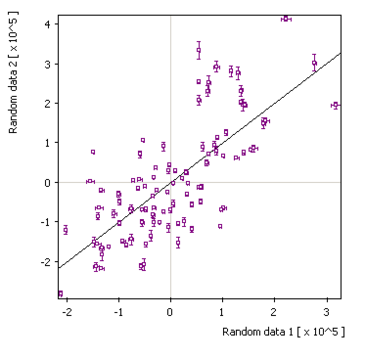
x/y + error: |
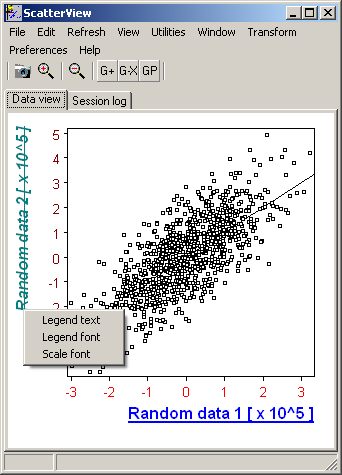
Symbols: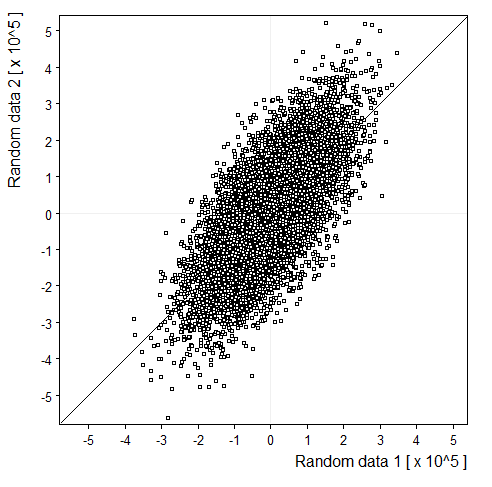 |
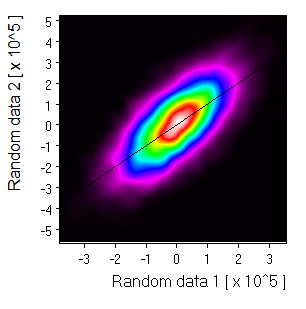
Density map: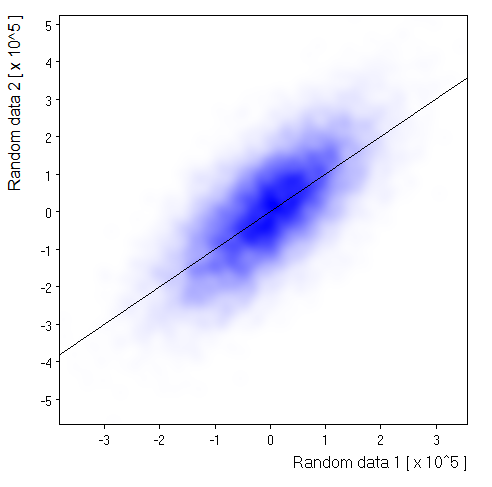 |
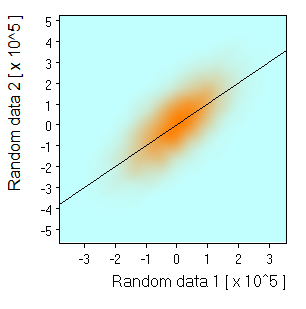
Monochrome: |
2-Color: |
3-color: |
Direct line: |
Right step: |
Left step: |
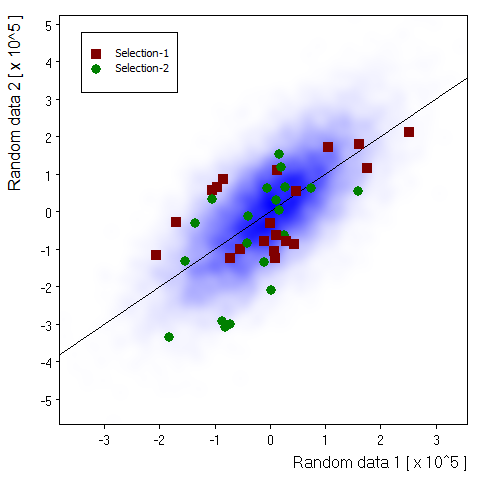
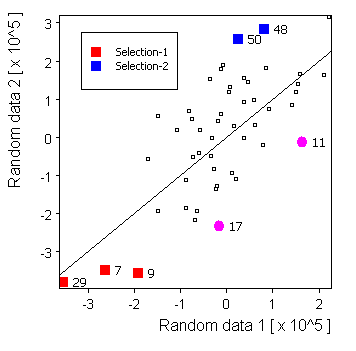
Selected source: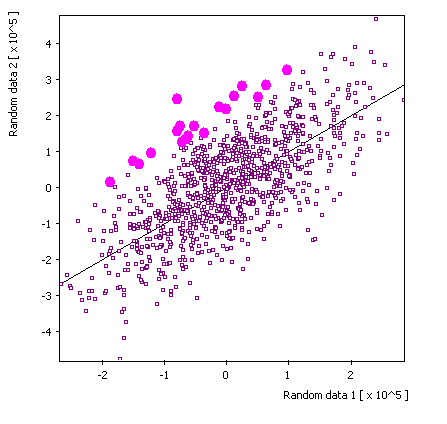 |
Replica 2: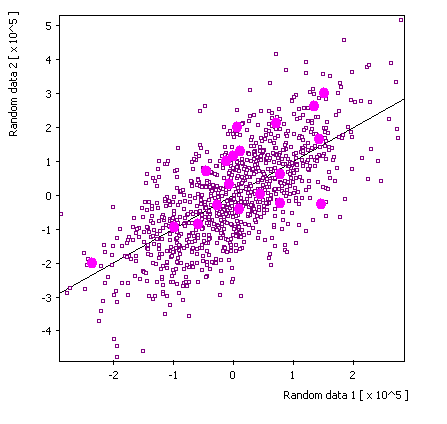 |
Replica 3: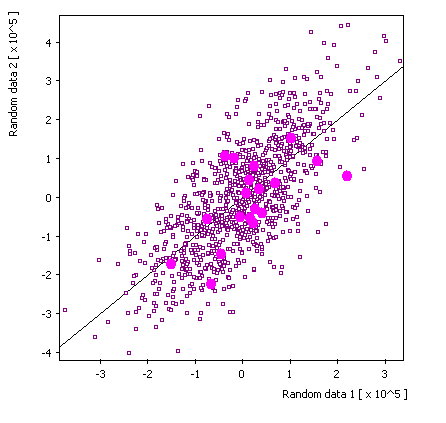 |
Linear: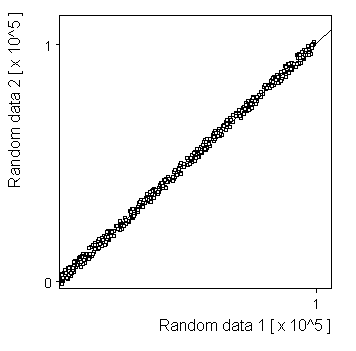 |
Gaussian: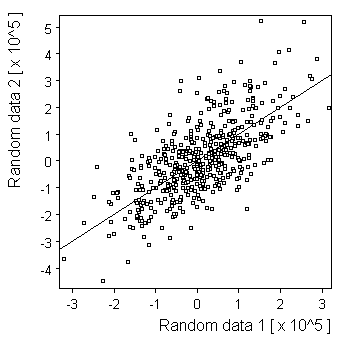 |
Half circle: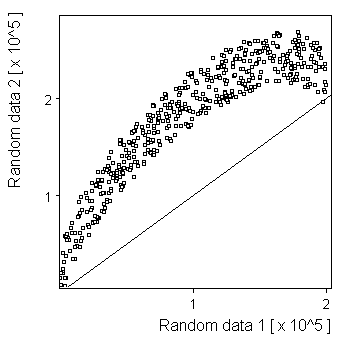 |
Waves: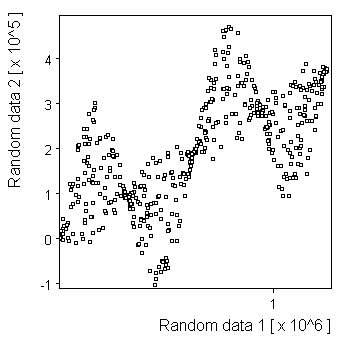 |
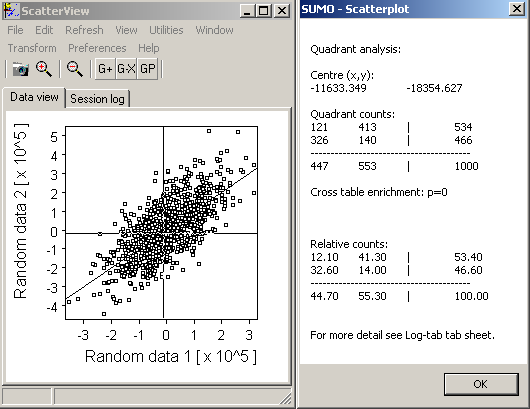
| Size=11,Sdev=2,Linear: | 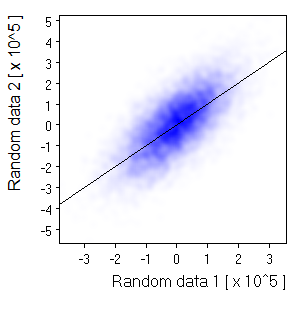 | |
| Size=21,Sdev=4,Linear: | 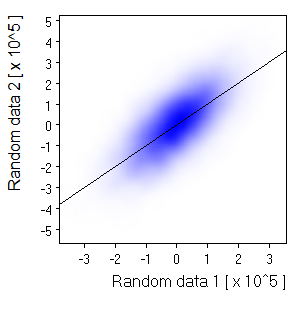 Nice and smooth for a small picture. But not if you expand the image | |
| Size=91,Sdev=10,Linear: | 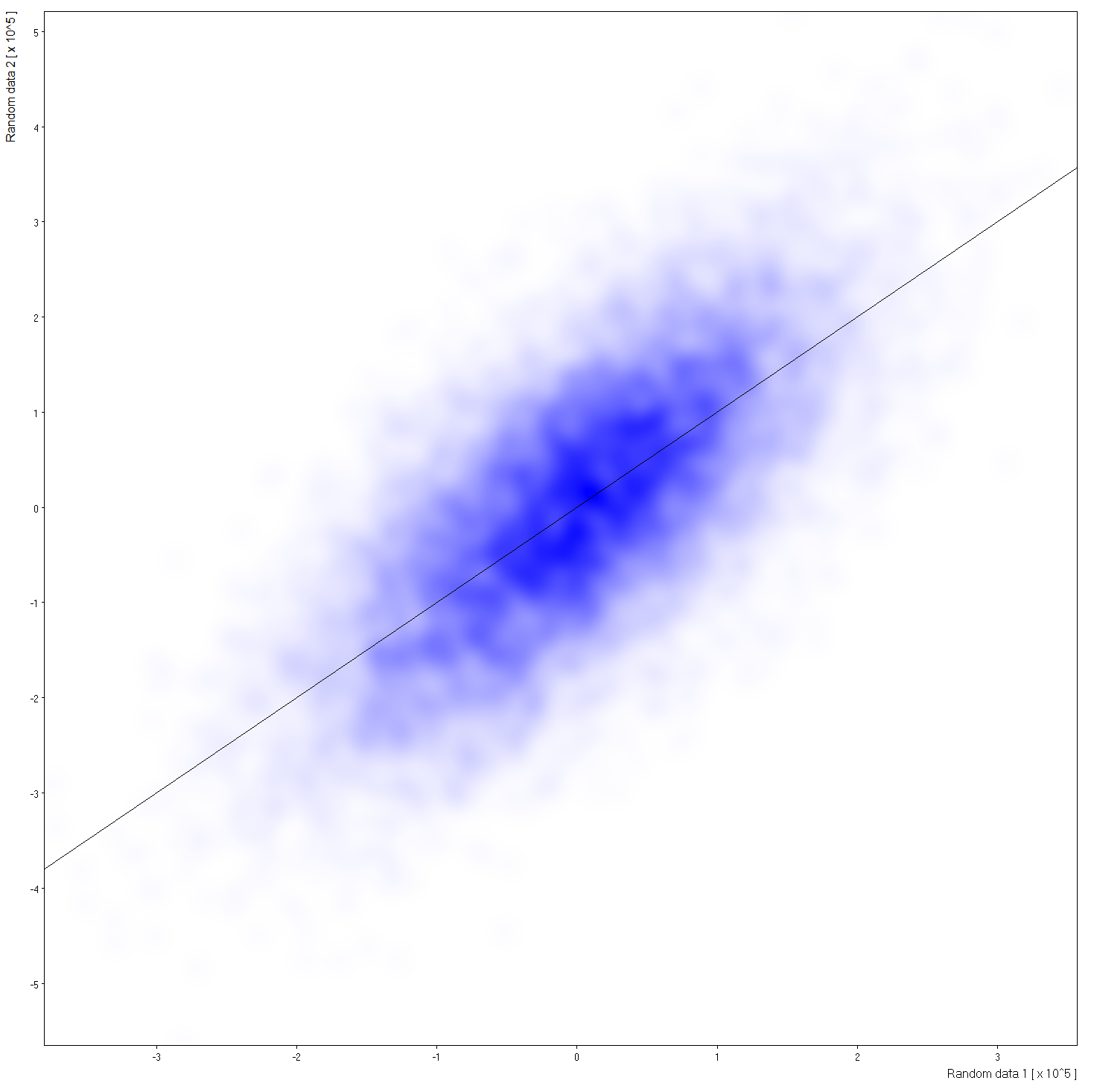 Click to see large view. | |
| Size=21,Sdev=7,Linear: | 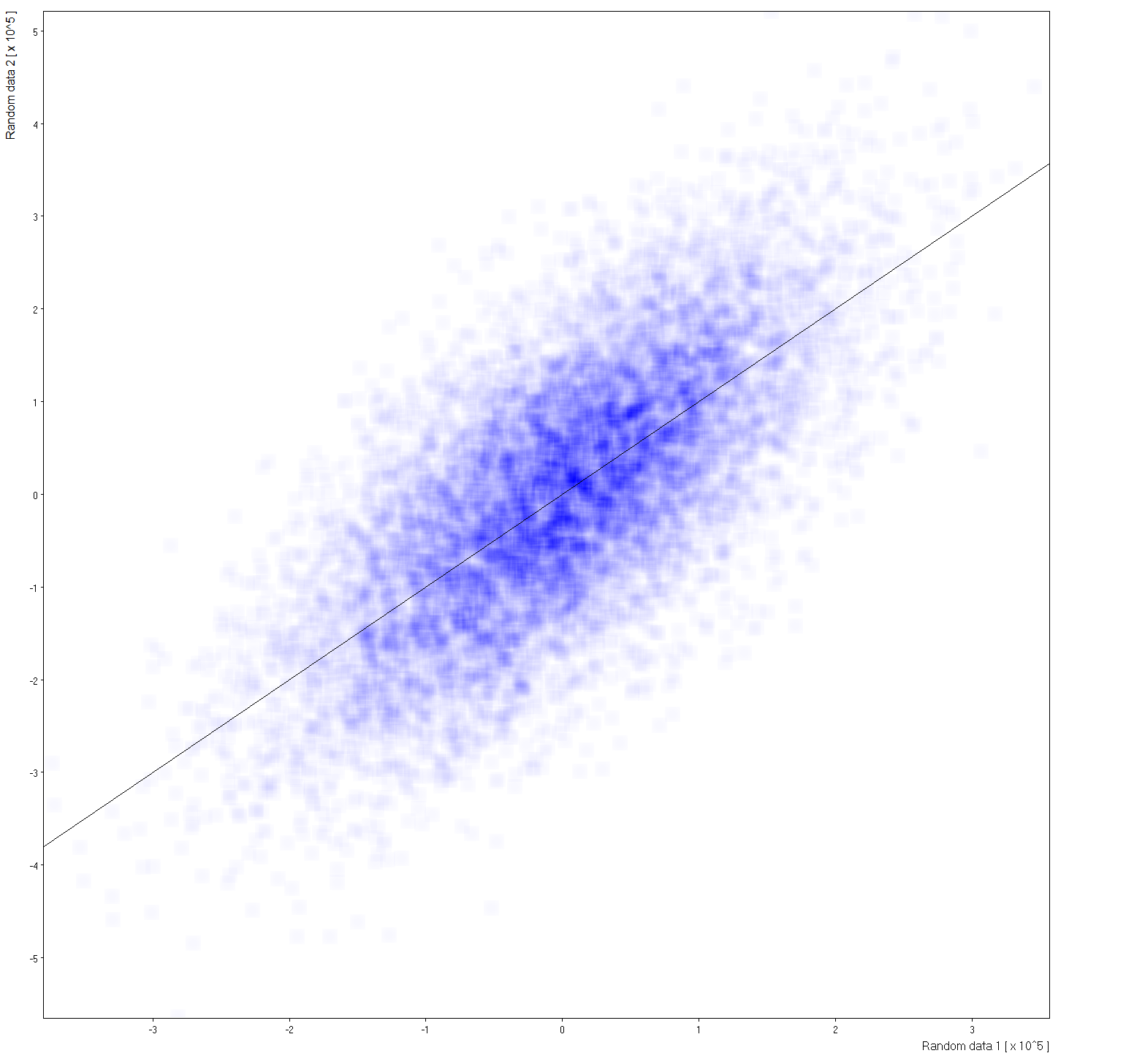 Gaussan profile to wide for computation kernel. Click to see large view. |
White=>Blue |
White=>Red |
White=>Green |