Gene-Network Builder 
Statistical tests generate lists of significantly (more or less) regulated genes.
They do not tell us anything about biological (medical) importance or impact.
One way to explore this could be, to use already existing informations about interactions
between genes / proteins /... .
Basic idea
NetBuilder
Load data
Data view
Preferences
Search nodes
Copy selected nodes
Paste a list of search keys
Data files
HPRD
NCBI
Gene Index
Network viewer
Basic Idea
A variaty of expermiments have been performed in the scientific community to explore which genes
/ proteins interact directly with other genes / proteins.
E.g. the Yeast.-2-Hybrid system has been used
to systematically explore direct binary interactions between proteins or between proteins and DNA.
As result a list of binary protein interactions is generated.
Assume a (partial) interaction list (data base) like:
| Interactor1 |
Interactor2 |
| P1 |
P450 |
| P2 |
P1730 |
| P15 |
P1 |
| P37 |
P260 |
| P165 |
P7612 |
| P450 |
P2917 |
| P765 |
P413 |
| P1216P |
P260 |
| P2917 |
P15 |
| P4579 |
P1213 |
| P5981 |
P37 |
| P7192 |
PP2917 |
| P9365 |
P4376 |
| ... |
... |
Assume your statistical test / cluster analysis results in a list of significant genes:
| P7,P15,P32,P176,P1875,P2917,P9123,... |
Now you can search the members of your gene list in the interaction database:
P7,P15,P32,P165,P1875,P2917,
P7192,P9123, ...
| Interactor1 |
Interactor2 |
| P1 |
P450 |
| P2 |
P1730 |
|
>P1 |
| P15 |
P260 |
| P165 |
P7612 |
| P450 |
P2917 |
| P765 |
P413 |
| P1216P |
P260 |
| P2917 |
P15 |
| P4579 |
P1213 |
| P5981 |
P37 |
| P7192 |
P2917 |
| P9123 |
P4376 |
| ... |
... |
Genes P7,P32,P1875 are not found at all in the interaction list => they can not be used for building of
gene-networks.
Genes P15,P165, P2917,P7192,
P9123 are found in the interaction database => they can be used for building of networks.
Direct interaction network:
In the most simple case we can try to build direct interaction networks: We onyl use
interaction partners which are also part of our list of significant genes.:
Lets look on our genes:
P15: partners P1, P260 - but these interactors are not
found in our list of significant genes, P2917 - member of our list.
P165 : partner P7612 no member.
P2917 : partner P450 - no member, P15, P7192 - member of our list.
P7192 : P2917 - member of our list.
P9123 : partner P4376 - no member
Thus we can try to build a networks with genes P15,P2917,P7192, resulting in a simple linear network:
The other two genes (P165, P9123 )
have interactions partners, but those are not members of our gene list => we don't use them.
One Gene Interpolation network
Assume the following genes:
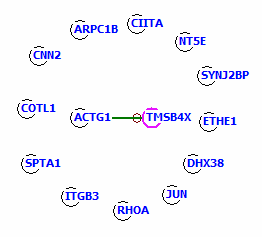
Only the ACTG1 and TMSB4X genes are direct interaction partner.
But we can try to interpolate genes. I.e. we try to find genes from our interaction database which might be
linking bridges between our selection genes.
Limitation: there should be only one linking gene.
E.g. Selection gene - Interpolation gene - Selection gene.
Bridges with multiple linking genes are not considered (e.g. Selection - Interpolate - Interpolate - Selection).
With this approach we could improve the above network:
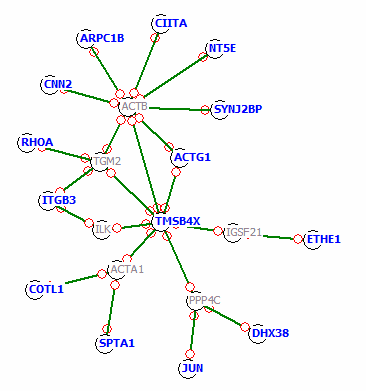
The light gray genes ACTB, TGM2, ILK, ACTA1, PPP4c and IGSF21 were interpolated.
I.e. they were NOT in our selection list - we did not identify them to be e.g. differentially regulated in our experiment.
But with the help of these 6 interpolated genes we can place all the other selection genes in one network - and possibly into one
biological context.
To switch between the two network modes, click the tool-bar buttons:
 |
Click the button to select building of Direct
Interaction networks (Default setting) |
 |
Click the button to select building of One-Gene-Interpolation
networks |
Evidence for a network
Often the question arises: is my network specific or would you generate such kind of network with any arbitrarily selected set of genes.
One way to answer this questions could be to test this with random gene-lists.
SUMO allows to generate a user defined number of random genelists with given size, and try to build nets.
A histogram ist build giving the number of nets with a certain numer of nodes:
Build random nets
======================
V1.00a, 18.05.2015; c.schwager@dkfz.de
Number of interacting genes: 16242
Number of interactions: 345992
Size of geneset: 100
Number of permutations: 100
Total number of nets found= 340
4.399s elapsed time.
Size distribution of random Nets
Size n C-Sum p
67 1 1 0.0029411765281111
15 1 2 0.0058823530562222
13 2 4 0.0117647061124444
12 4 8 0.0235294122248888
11 1 9 0.0264705885201693
9 2 11 0.0323529429733753
8 5 16 0.0470588244497776
7 4 20 0.0588235296308994
6 4 24 0.0705882385373116
5 7 31 0.0911764726042747
4 12 43 0.126470595598221
3 61 104 0.30588236451149
2 236 340 1
The table sumarizes the permutation analysis results:
| Size | Size of of the net = number of nodes in the net |
| n | Number of nets with given size |
| C-Sum | Cumulative sum, the number of nets with given or evene larger size |
| p | probability values to find this (or larger sized networks |
This procedure does not explicitly take into account connectivity of single nodes.
E.g. a set of 5 linearly connected nodes will be treated exactly as a star configuration where any node connects any other:
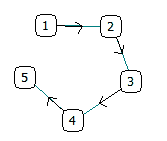 | = | 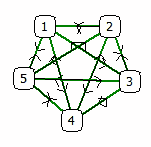 |
Obviously, the p-value depends on the size of the genelist as well as on the interaction data base (number of genes and interactions).
Thus, you can estimate the probaility of a certain sized net to be generated by random genelists.
In SUMO Select Netbuilder | Utilities | Random net (to test for user defined genelist size), or
select "P-permutation" from context menu in the analysis tree for a specific net.
NetBuilder
In SUMO click Gene-Net Builder button, or select NetBuilder
from the analysis menu:
An empty Net-Builder windows opens:
Ensure databases are loaded.
Use Preferences tab to set-up database parameters.
Next paste a genelist from clipboard or load a gene-list file.
Paste a genelist
Select Edit | Paste genelist from main menu to to get a list of genes from
clipboard for network analysis.
At present, SUMO expects a list of gene symbols (e.g RAS, MYC, HPRT1, VEGFR, ....)
Genes should be separated by spaces, tabs, commas, semi colons or one gene per line.
You can also supply regulation information for each pasted gene.
In this case NetBuilder expects one gene per line followed by a
positive or negative number, sperated by a tab (e.g. VEGF tab
-0.78).
NetBuilder automatically replaces German decimal comma by English
decimal point.
Genes are compared with the loaded interactions, and a Direct-Interaction network (see above:
Basic Idea) is built.
In the analysis tree a new noded is added:
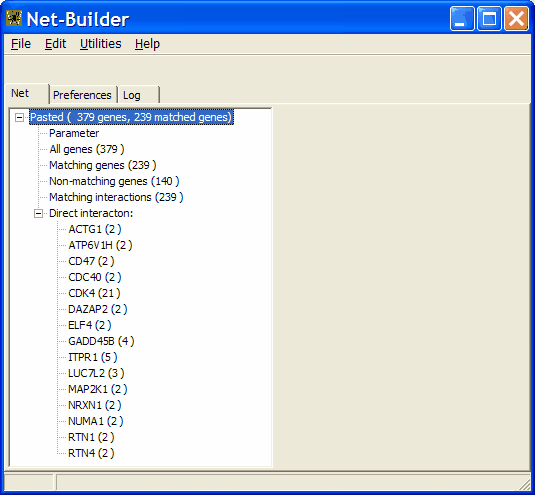
You can see basic statistics
- Number of pasted gene names (All genes 379)
- Number of genes mapped on in interactions ( Matching genes 239)
- Number of genes not found in intgeraction data base (Non-matching genes 140)
- assembled direct interaction networks, including number of genes wihtin each single net.<
Click a gene list (e.g. Non-Matching genes). The corresponding genes are listed in
Log-tabsheet and may be copied.
Click a net (e.g. CDK4 (21) ). The net is shown in a basic network viewer:
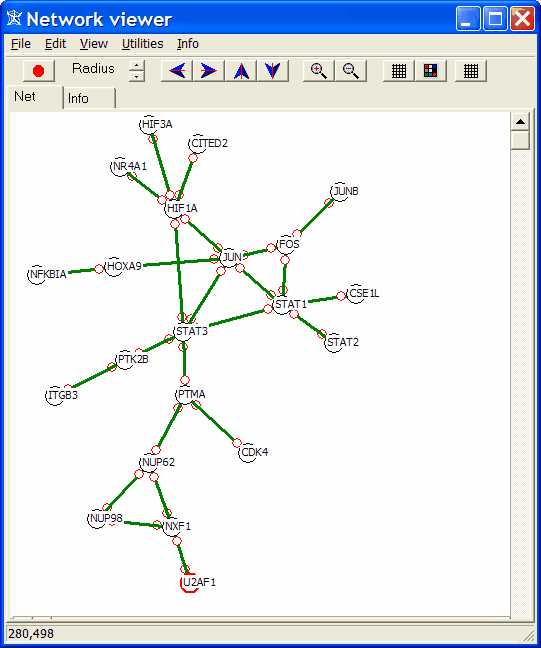
You can rearrange single (or multiple nodes) by selecting and freely dragging them.
Preferences
Here you can customize Net-Builder.
Most important: Define interaction data-bases and gene index.
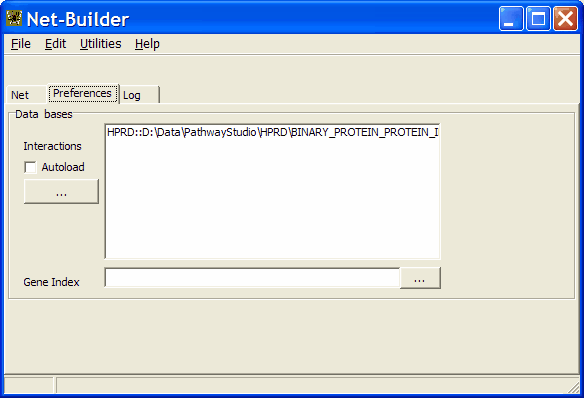
In the data bases field you can define the available interaction data-base files.
To add a new one click the " ... " button.
Select the data base type (HPRD or NCBI). Next select a suited file.
In the example a single data base file is defined. It contains all direct binary protein-protein interactions availabe from
HPRD.
In this case the data base identifier is: HPRD::
followed by the file specification: D:\Data\...
See the presently supported data-bases.
To load a defined interaction data base double-click the corresponding line in the list.
Check the Autoload checkbox:
Next time you open Net-Builder the defined interacton databases are loaded automatically.
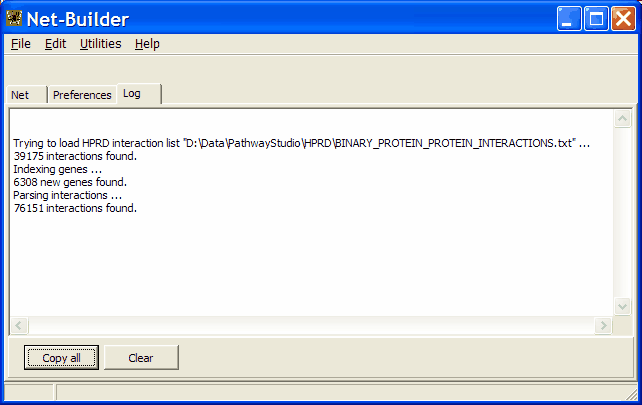
Review the Log-tabsheet to see the data-base loading success:
Presently supported data-base formats:
HPRD::
Human Protein Reference data base.
From their download site you can download a list of binary protein-protein
interactions.
Download the file interactions.gz and unpack the contained interaction list (
Use the file "... containing human binary protein-protein
interactions in tab delimited format."
Each line in the data file describes a binary interaction, e.g.:
PAG1
05772 NP_060910.3 VAV1 01284
NP_005419.2 in vitro 10790433
| interactor_1_geneSymbol |
PAG1 |
| interactor_1_hprd_id |
05772 |
| interactor_1_refseq_id |
NP_060910.3 |
| interactor_2_geneSymbol |
VAV1 |
| interactor_2_hprd_id |
01284 |
| interactor_2_refseq_id |
NP_005419.2 |
| experiment_type |
in vitro |
| reference_id |
10790433 |
At present, Net-Builder only uses the Gene symbol.
Any other interaction list may be easily converted into a format compatible to the HPRD format and loaded into Net-Builder.
NCBI::
NCBI's interaction database.
From their ftp site you can download a list of binary protein-protein
interactions. Presently, the NCBI list contains interactions from three sources: HPRD, BIND and BIOGRID.
Download the interactons.gz file
and unpack the interaction list with a suited unzip program (e.g. 










