Sometimes your data matrix may contain non
numerical values. (e.g. NAN = Not A Number as result of illegal mathematical
upstream operations in the data pre-processing; or any other text which can not
be converted into a floating point number).
SUMO converts these data cells into "Zero", which may add a
BIAS to your data and subsequent statistical analyses.
It might be be better to impute those missing values in a well defined manner by
more or less intelligent estimated values.
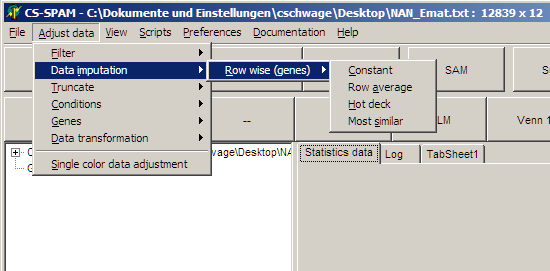
SUMO offers four methods for data imputation:
Constant : all missing values are replaced by a user defined constant:
Row average : The average of all data values (excluding the missing values) is calculated. This average is used to replace the missing values.
Hot deck : An arbitrary value from the genes other data values is used to replaced the missing value.
Most similar : Most similar genes are searched (euclidean distance, excluding the positions where missing values were found). From those an average for the missing values is calculated and inserted.
SUMO can impute multiple missing values within a gene in one go. Already imputed values are not influencing the other genes (non-recursive imputation).
Select Main menu | Adjust data | Data
imputation | Row wise

The group selection dialog pops-up:
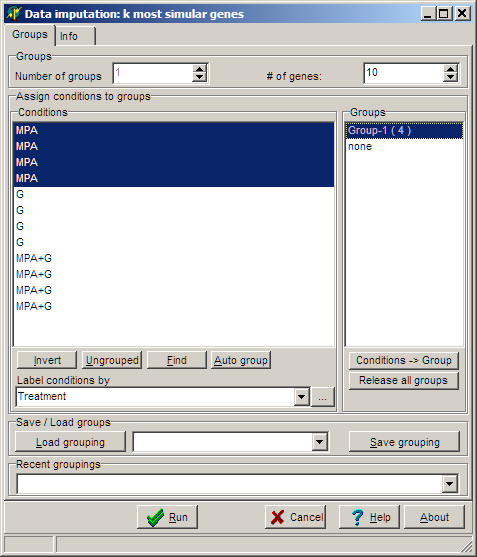
In the example we first
try to impute missing values filter for the "MPA" hybs, then the "G" hybs and finally the "MPA+G" hybs.
With the # of genes edit field you can define how many most similar genes
are search and averaged.
!! ----- NB ----- !!
Imputations should only be performed for a small number of missing values (10%
of a group or less).
Otherwise a strong BIAS is added to the data resulting in misleading statistical
analyses.
Use the filter to
remove genes with too many missing values.
last edited 23.09.2007