Some times it is required to re-calculate data columns in hybridzation data files :
Such functionality is found in the Compute tabsheet.
All operations are performed on single rows, sometimes multiple or even
the whole data file are used to calculate data values which will be placed in a
single row.
If you want to recompute columns goto to the Column CPU tabsheet.
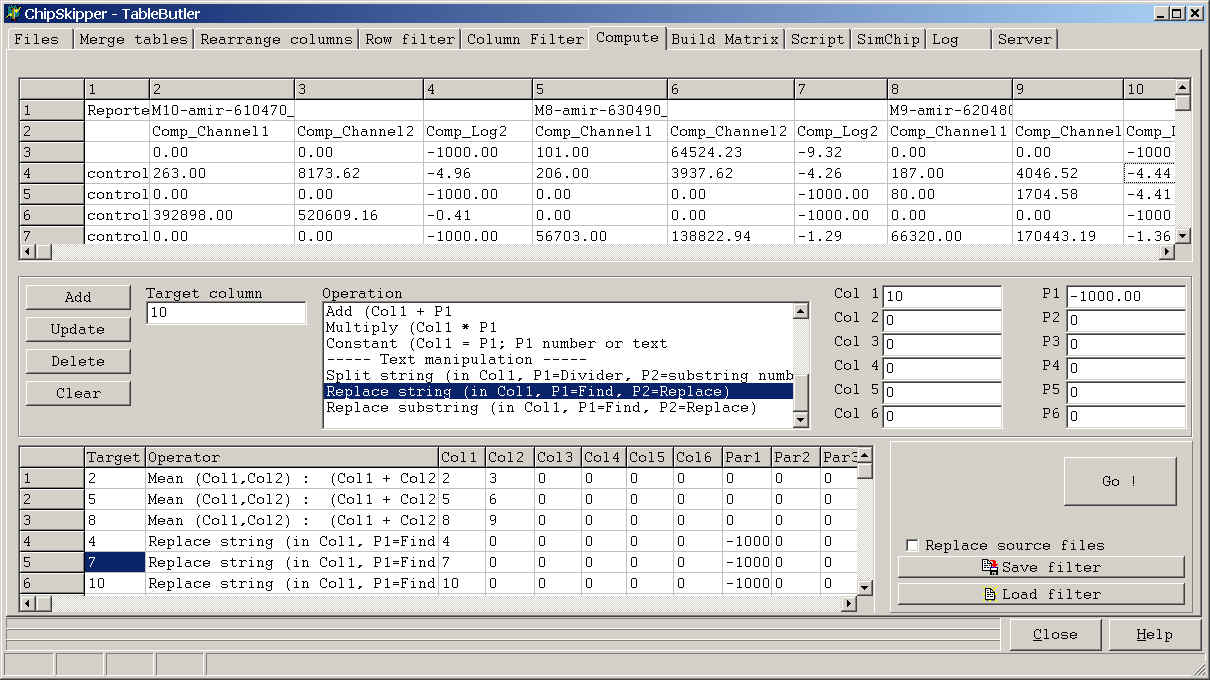
TableButler shows a preview of the data file :

To set-up a single Operation do the following steps:
By default, TableButler save the result in a new file with extension "_cpu" (e.g.
selected file is "my_table.txt", a result file with name "my_table.txt_cpu" wil
be created.
To overwrite the source file check the Replace Source check box.
If you want to reuse a set-up list of operations, save the "filter" by
clicking the Save filter button.
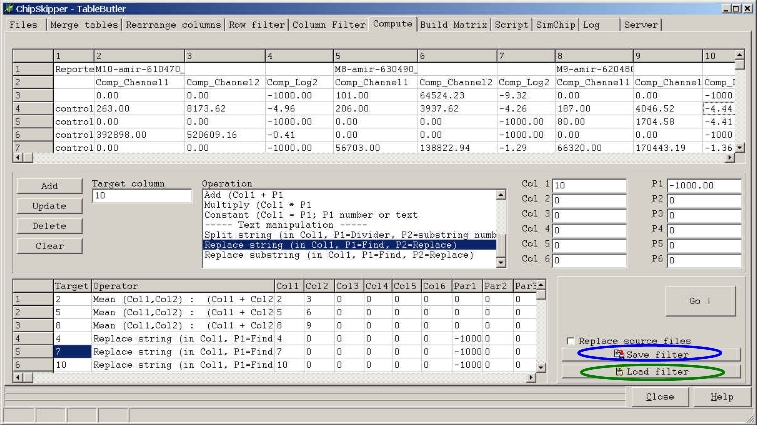
In a next session reload the filter by clicking the Load filter button.
Click a colum in the preview grid, then click the Target column field to define where the result of the operation shold be placed, or leave target column empty tp append the new data as new data fields.
If you later click the Add button the respective Target Column value will
show up in the Filters list.
If you do not supply a Target Column value (= leave field empty) or type
new, the result of the operation will be appended to the table.
Some operations will create multiple column results or place the results in the
source cells.
See more details for the respective functions.
Click a colum in the preview grid, then click the
Col1, Col2,
...
to select the columns for the operation:
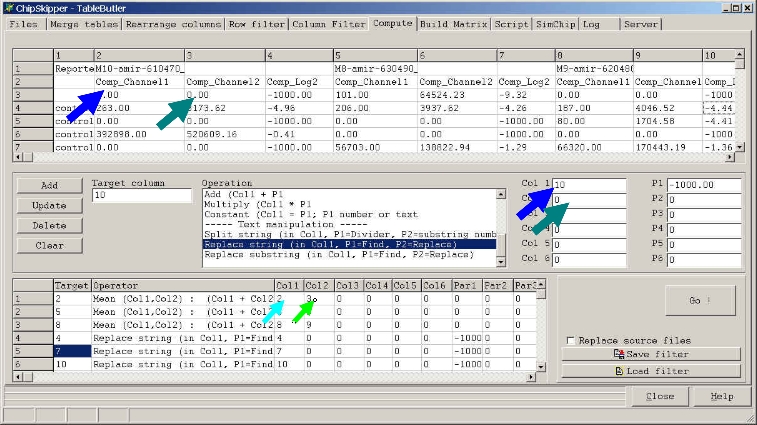
If you later click the Add button the respective Target Column value will show
up in the Filters list.
If you want / need to define a larger number of source columns you can also
define a simple regular expression range expression in edit field Col1.
Either define a
e.g. 2,3,6..10,12,13,20..24 will select columns 2,3,6,7,8,9,10,12,13,20,21,22,23,24
You can also use add multiple coluns to the Col1-edit field by Double-Clicking a column in the preview grid while holdeing Ctrl-Key pressed.
Select the requested operation from the Operation list:

To get a brief description pf the selected operation, click the
information button ( "???" ).
A windows message box pops-up showing brief essential information about
the selected operation.

For more details about a operation see below.
At present the following groups and operations are available:
Basic arithmetics :
Constant value is defined in edit field P1
Data adjustment
Basic Statistics
Divisive statistics
Row arithmetics
Conditional operations
Text operations
Coordinate transformation
Enter required parameters into P1..P6
Add filter to the list of filters and run filter.
A defined filter may contain multiple operations and may be saved for later use.
The examples shows recalculation of combined data files created with Expression matrix builder of TableButler data file :
1..3 : Calc Average Intensity from Red and Green signal2. Spot should belong
to the 75% brightest spots in Green color channel
4..6 : Replace "-1000.00" expression values from empty spots with
"0".
This function splits the contents of a cell into
individual cells. Target cells are adressed by tags found in the original cell.
TableButler can handle two cases:
Example1:
assume you have a gene annotation column like:
| Header line | GeneAnno |
| ... | ... |
| Gene25: | [Genbank]123498;[ENSEMBLE]AS34985 |
| ... | ... |
| Gene77: | [Locuslink]234567;[GO]456789 |
| ... | ... |
| Gene88: | [Genbank]678990];[Swissprot]788901 |
| ... | ... |
TableButler will generate new column with header names
[Genbank] [ENSEMBLE] [Locuslink] [GO] [Swissprot]
and sort the corresponding "values" of all genes sorted into the respective columns:
| Header line | Genbank | ENSEMBLE | Locuslink | GO | Swissprot |
| ... | ... | ||||
| Gene25: | 123498 | AS34985 | -- | -- | -- |
| ... | ... | ||||
| Gene77: | -- | -- | 234567 | 456789 | -- |
| ... | ... | ||||
| Gene88: | 678990 | -- | -- | -- | 788901 |
| ... | ... |
The number of inserted columns is
dependant of the found tags.
Therefore 'this function should be run INDEPENDANT and NOT in combination with
other functions where Column addresses are required.
Parameters:
Col1 edit filed = ID of column to split
P1 edit field = character (or string) which divides
single fields.
In the example ";"
P2 edit field = character or string wich divides "Tag"
from "Value"
In the the example "]"
Example2:
assume you have a gene annotations like:
| Header line | GeneAnno |
| ... | ... |
| Gene25: | Genbank|123498|ENSEMBLE|AS34985 |
| ... | ... |
| Gene77: | Locuslink|234567|GO]456789 |
| ... | ... |
| Gene88: | Genbank|678990|Swissprot|788901 |
| ... | ... |
Parameters:
Col1 = as above
P1= the onyl field divider
In the example "|"
P2 edit filed MUST be empty
!!!!!!!
Any value in P2 would be interpreted as divider and thus scramble the operation.
Assume a result file from micro array scanning where for unique genes there
were muliple replicates on the microarray. Furthermore you have different slide
layouts where the total number of features and the number of replicas per
feature are different.
Trying to merge such kind of data on a per feature basis is problematic.
Here it might be better to first average replicas per single slide layout (or
even per single hyb) and then join the differtn layouts using UNIQUE gene-IDs.
Replicate averaging allows to do this.
Define the column which contains the Key (e.g. gene name) used to find
replicas in TARGET COLUMN edit field.
Define the columns for which replica averages shall be calculated in edit
fields (COL1 ..COL12). At present only 12 columns may be averaged in one run.
A result file is created which contains
Key-Column, Averages Col1..Col12, Number of replicas:
| ID | Arithmetic_Mean | Geometric_Mean | # |
| A_23_P101972 | 55.18 | 51.78 | 1 |
| A_23_P101992 | 42.61 | 35.97 | 1 |
| A_23_P102000 | 816.484 | 92.21 | 10 |
| A_23_P102017 | 8.59 | 5.96 | 1 |
| A_23_P102037 | 274.42 | 269 | 1 |
| A_23_P102058 | 72.69 | 52 | 1 |
| A_23_P102454 | 779.68 | 728.34 | 1 |
| A_23_P102462 | 118.35 | 110.66 | 1 |
| A_23_P102471 | 2315.386 | 2136.46 | 10 |
| A_23_P102481 | 12.25 | 8.68 | 1 |
| A_23_P102494 | 326.35 | 302.44 | 1 |
Last edited 22.11.2007,